Merge branch 'enable_go' into 'dev'
Enable go See merge request !17
No related branches found
No related tags found
This commit is part of merge request !18. Comments created here will be created in the context of that merge request.
Showing
- README.md 18 additions, 2 deletionsREADME.md
- constants.py 1 addition, 0 deletionsconstants.py
- examples/citrus_sinensis.yml 4 additions, 4 deletionsexamples/citrus_sinensis.yml
- gga_init.py 1 addition, 1 deletiongga_init.py
- images/gga_schema.png 0 additions, 0 deletionsimages/gga_schema.png
- speciesData.py 9 additions, 5 deletionsspeciesData.py
- templates/gspecies_compose.yml.j2 1 addition, 1 deletiontemplates/gspecies_compose.yml.j2
- templates/organisms.yml.j2 2 additions, 1 deletiontemplates/organisms.yml.j2
- utilities.py 1 addition, 0 deletionsutilities.py
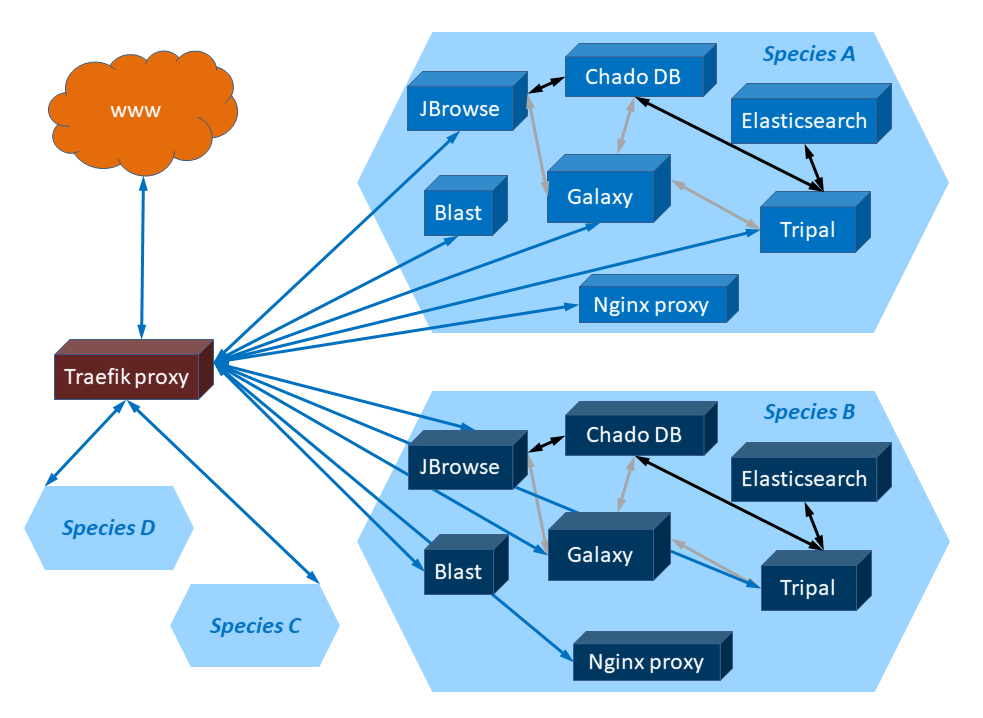
images/gga_schema.png
0 → 100644
113 KiB